Colabrativ’s work with Biogen Idec Inc. on the structural comparison of Factor VIII (FVIII) and recombinant Factor VIII-Fc fusion protein (rFVIIIFc) using Small-Angle X-ray Scattering (SAXS) was presented at the 54th Annual Meeting of the American Society of Hematology on December 8, 2012. The presentation also included results of a hydrogen/deuterium exchange (H/DX) mass spectrometry comparison of the two proteins from the Protein Formulation Development group at Biogen Idec which was in agreement with the SAXS data. The title of the presentation was “Structural Comparability between Recombinant FVIII-Fc and its Isolated FVIII and Fc Constituents” and the abstract can be found at https://ash.confex.com/ash/2012/webprogram/Paper53949.html.
Colabrativ’s Role in Small-Angle X-ray Scattering Experiments
Colabrativ’s role in this work started at the conceptual level during discussions with the Biogen Idec scientists about structural approaches to compare the FVIII and rFVIIIFc proteins. Crystallization was an obvious choice, but due to the flexible linker between the FVIII and the Fc in rFVIIIFc, crystallization of the rFVIIIFc was considered to have a low probability of success. SAXS had it own challenges, i.e. obtaining aggregate free samples of both proteins at reasonable concentrations. Early on while evaluating SAXS as an analysis option, we contacted Dr. John Tainer, who put us in touch with Dr. Susan Tsutakawa at SIBYLS beamline 12.3.1 at the Advanced Light Source (ALS). We made the contractual arrangements with the ALS and worked with the Biogen Idec staff to prepare and submit a successful SAXS proposal. We worked with both the Biogen Idec and ALS staffs on the preparation and shipping of the protein to the ALS. Working with Dr. Tsutakawa, we purified both proteins at the ALS by size-exclusion chromatography, prepared in a range of concentrations, and collected the SAXS data on the samples and their corresponding filtrates all in the same day.
Analysis of Factor VIII and Factor VIII-Fc SAXS Data
The analysis of the SAXS data and model refinement was performed by Dr. Tsutakawa beginning with initial models supplied by Biogen Idec.
The Guinier plots of the SAXS data for FVIII and rFVIIIFc showed both samples to be monodisperse; i.e. they have linear Guinier plot slopes. The radii of gyration (Rg) for FVIII and rFVIIIFc from the Guinier plots are 38Å and 51Å, respectively. The Electron Pair Distributions have a Dmax of 125Å and 175Å for FVIII and rFVIIIFc, respectively.
Models of FVIII and rFVIIIFc used the crystallographic structures of Factor VIII (3CDZ.pdb) and Fc as starting models. The program BILBOMD was used to identify a small set of conformations for these starting coordinates that best match the experimental SAXS data. Missing loops and carbohydrate chains were added to improve the fit to the SAXS data. For FVIII, the Chi value improved from 3.5 to 1.8 after modeling the missing loops and optimizing the conformation with BILBOMD, and to 1.5 after modeling the missing carbohydrate structures; see Figure 1.
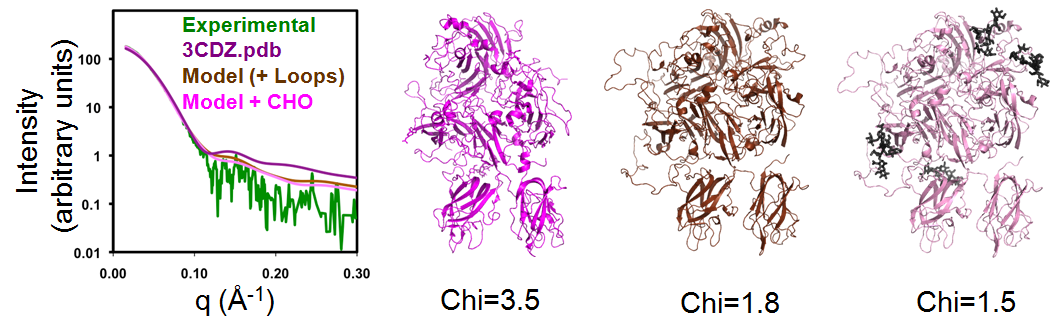
|
| The molecular dynamics simulation program BILBOMD was used to model conformers of BDD FVIII and compare their theoretical X-ray scattering curves to experimental scattering data. Three models are shown: The crystal structure of BDD FVIII (3CDZ.pdb – Chi=3.5), BDD FVIII with loops (Chi=1.8), and BDD FVIII with addition of N-linked carbohydrate (Chi=1.5). The fit of the three FVIII models to the experimental SAXS data is shown in the left panel. |
The rFVIIIFc BILBOMD model has two conformations contributing to the best fit of SAXS data when only the missing loops were added. Unlike the FVIII modeling, the fit to the SAXS data did not improve when the carbohydrate structures were modeled and optimized with BILBOMD; see Figure 2.
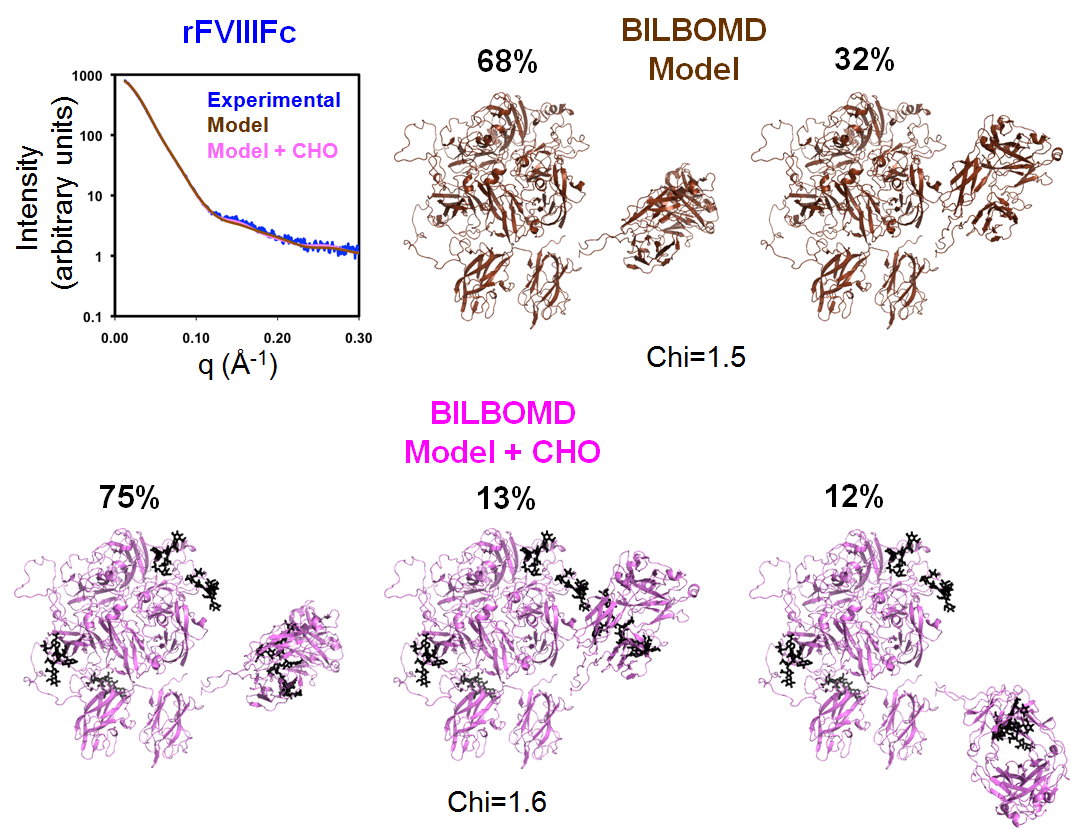
|
| BILBOMD was used to generate a minimal ensemble of structures for which theoretical X-ray scattering curves optimally fit experimental scattering data. Two models are shown: The rFVIIIFc model based on the BDD and Fc crystal structures with surface loops but lacking N glycans has a (Chi=1.5), and the same model with of N-linked carbohydrate present (Chi=1.6). The fit of the two rFVIIIFc models to the experimental SAXS data is shown in the upper-left panel. |
Conclusion
The Fc domain in rFVIIIFc extends away from the regions in FVIII that are essential for interaction with elements of the Xase complex (anionic phospholipids, Factor IXa, and Factor X) and von Willebrand factor. This result is consistent with the conclusions from the H/DX study that shows that the fusion of Fc to FVIII does not perturb the structure of either the FVIII or Fc elements of rFVIIIFc.

Thanks for the great article..